GOALS
While the ability of cells to communicate through the exchange of exRNA has now been confirmed across all domains of life, the mechanistic details regarding how exRNAs are selected for secretion, how these exRNA identify their target cells, and the effects their delivery have on cellular recipients remain largely undescribed.
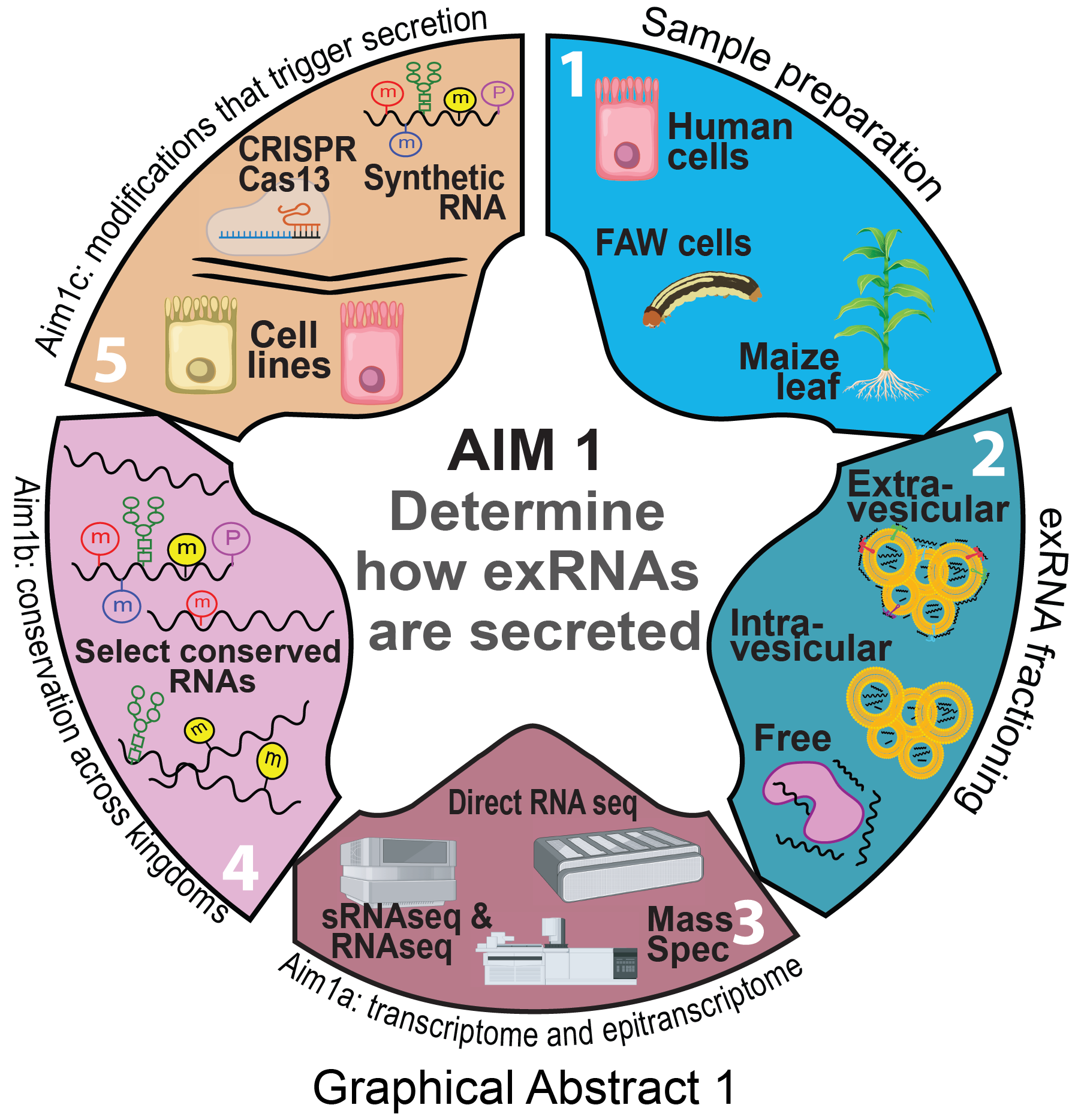
Aim 1: How are exRNAs targeted?
Hypothesis: exRNAs have specific post-transcriptional modifications and/or sequence characteristics (or referred to as ZIP codes) that mark RNAs for extracellular secretion.
Identify ZIP codes enriched in exRNAs.
Identify specific ZIP codes significantly associated with exRNAs across kingdoms
Directly determine if specific modifications are necessary and/or sufficient for exRNA secretion.
Aim 2: Determine how exRNAs are selected for intercellular and inter-species communication.
Hypothesis: In both intra-organismal and inter-species cellular communication, RNAs are transferred from the signaler cell (plant/insect/vertebrate) into the receiver cell (bacteria/self); and the ZIP codes used for transfer into bacteria differ from those used for transfer into self cells.
Characterize exRNAs participating in inter-species cellular communication
Characterize exRNAs participating in intra-organismal cellular communication
Define targets of individual signaler exRNAs in recipient cells.
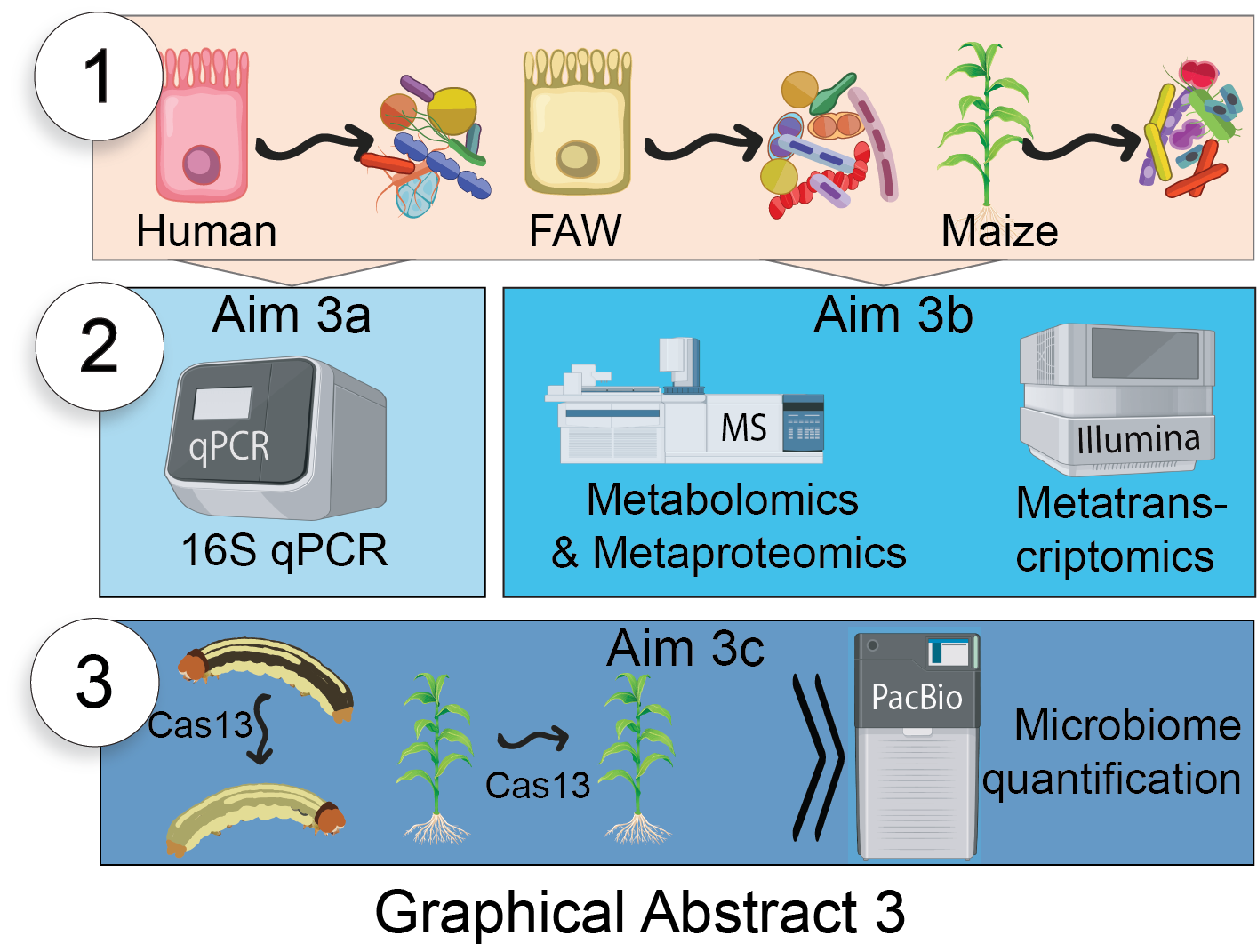
Aim 3: Determine the effects of eukaryotic exRNAs on associated microbiomes.
Hypothesis: exRNA-based communications is a direct mediator of host-microbe interactions critical to microbiome health and composition.
Determine the effects of exRNAs on bacterial community structure
Assess exRNA impact on bacterial metatranscriptomes, metaproteomes, and metabolites.
Test in vivo whether manipulation of specific exRNAs affects the microbiome community.